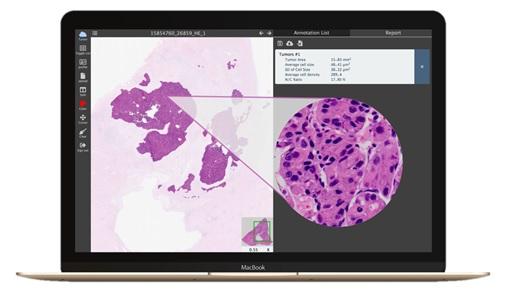
PC/Mac
Common tools for pathologists.

Our aspiration is to build the best A.I. analysis in pathology.
Our short term plan is to deliver various set of A.I. modules that allow pathologists to improve their workflow, and thus provide better service, at lower cost. Our long-term plan is to develop prognostic tools that integrate digital pathology with electronic health records, and other clinical data to provide pathologists with layers of information to better optimize patient's diagnosis.
Liver histopathology image analysis is a commonly used approach to confirm the decision in liver disease diagnosis. Current assistive histopathology image analysis systems have their function limitations in extracting morphology features, effective discrimination of cancer and normal tissues. Furthermore, some disease types of cells have highly varied features and ambiguous boundary. Some region segmentations such as the portal area segemention, also need to refer to surrounding cells. All of these characteristics make the analysis of liver histopathology images very challenging. In view of these limitations, we aims to develop new deep learning modules, combined with image analysis, for the analysis of liver histopathology images, the detection of liver cancers, and the grading of inflammation degrees. The results will be realized into a practical system for doctors to test and provide feedback, for achieving precision medicine.
We had build a cross-platform Giga-pixel viewer with cloud-architecture and to deliver an open workflow for pathologists. In current scenario, pathologists are limited to use the scanner and software provided by the vendor to digitize slides and analyze them. ALOVAS tackles the bindings and provides multiple A.I. algorithms to help pathologists contribute reports. The results are masked on the WSI digital slide and can be un-shown.
ALOVAS provides Real-Time magnification up to 40X and support 4 tabs at a time for multi-tasking, e.g. comparing different slides on a patient. Touch as input is supported, ALOVAS is supporting touch device such as Ipad and apple pencil. With supporting apple pencil, pathologists can use it as input to label and mark annotations smooth and natural.

Common tools for pathologists.
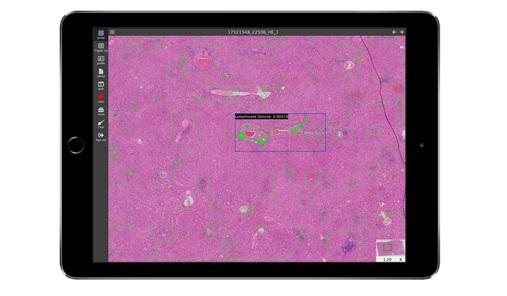
Support for apple pencil as input.
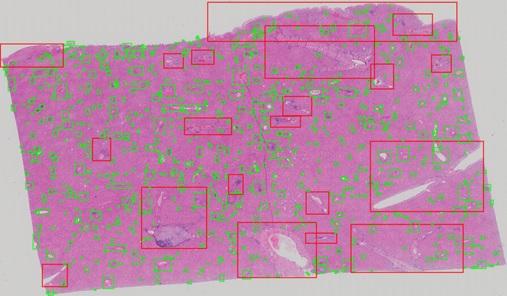
A multi-cscale convolutional neural network is utilized for patch-wise recognition of liver tumor with a detection performance of 95% mIOU. After the tumor is identified, the area is fed into another classifier for grading liver tissue as one of four different tumor grades. It combines a traditional image-processing based extractor retrieving sinusoid, cell and trabecular features as well as an CNN based extractor retrieving tissue features to achieve 89% grading accuracy.

Lymphocyters with different degrees of inflammation have different appearances, and they can be easily misjudged as epithelial cells and kuffer cells. A convolution layer of step-size 2 is proposed replace max-pooling to avoid feature loss in a U-net. The resulting U-net is used for automatic lymphocyte detection.
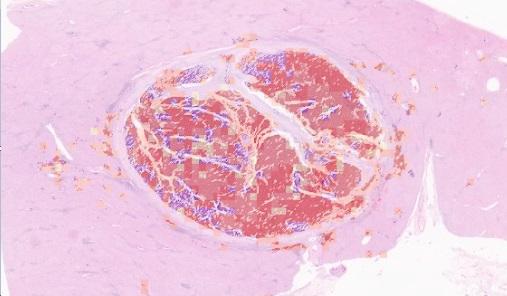
Segmenting portal areas can support hepatitis grading, as they are important regions to observe. An algorithm incorporating state-of-art CNN with multi-scale reference to liver portal is proposed to identify portal areas of various sizes and shapes.
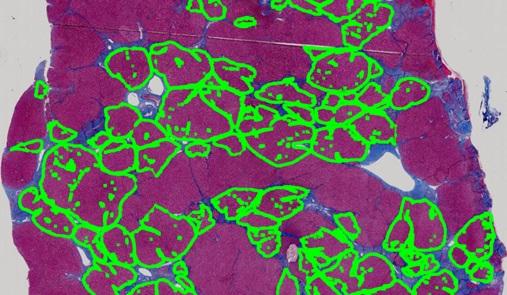
A framework is proposed to automate the calculation of Ishak Score for quantifying the degree of liver fibrosis. Ambiguous descriptions from human judgement can be therefore avoided in hepatitis diagnosis.
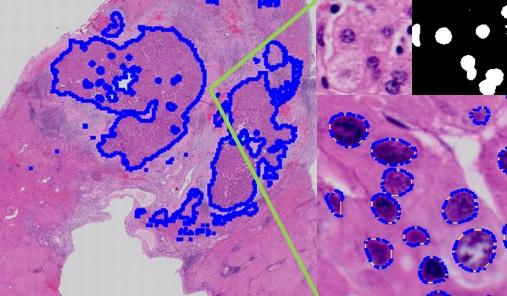
An algorithm is designed to provide quick nucleus segmentation with CNNs in given regions. The result then can be used to calculate medical parameters such as average size, density, standard deviation and N/C ratio of nuclei for analysis and diagnosis.
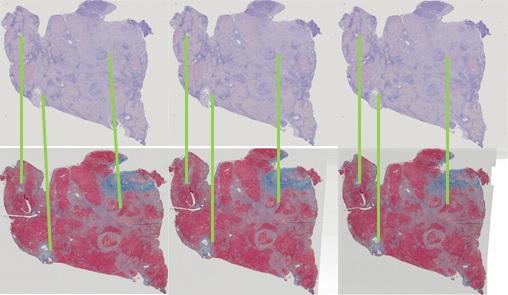
When one wants to observe various properties of the same region in multi-stained slides, an image registration process is required. The reason is that the slides are actually different slices so the cells' distributions are different. Additionally, the stained slides may suffer rotation and translation when scanned. An algorithm is proposed to find the best transformation parameters to align one slide with another.